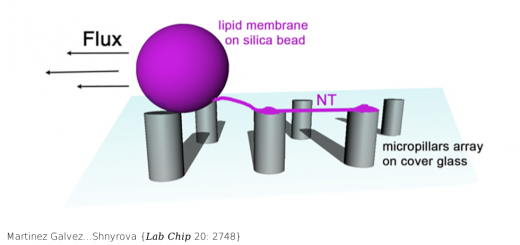
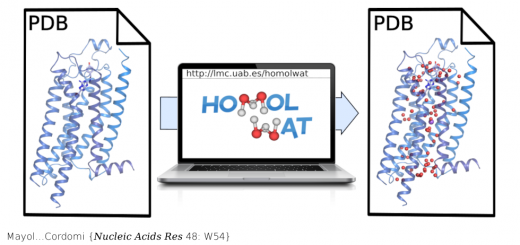
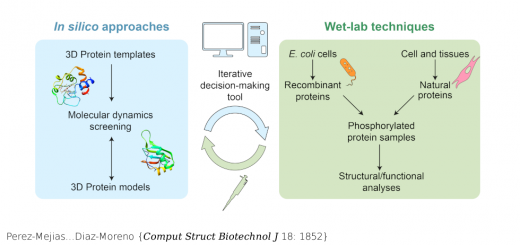
Terrazas…Orozco {Nucleic Acids Res 44: 4354}

In silico designed N-alkyl-N capped dumbbell-shaped structures were efficiently synthesized by double ligation of BCn-loop hairpins (n = number of carbon atoms of the alkyl chain). The resulting BCn-loop dumbbells displayed strong biostability and efficient RNA interference activity against a range of mRNA targets. The best dumbbell design (containing BC6 loops and 29 bp) was successfully used to silence GRB7 expression in HER2+ breast cancer cells for longer periods of time than natural siRNAs and known biostable dumbbells.
Rational design of novel N-alkyl-N capped biostable RNA nanostructures for efficient long-term inhibition of gene expression
Terrazas M, Ivani I, Villegas N, Paris C, Salvans C, Brun-Heath I, Orozco M.
Nucleic Acids Res 2016 May; 44: 4354.
Computational techniques have been used to design a novel class of RNA architecture with expected improved resistance to nuclease degradation, while showing interference RNA activity. The in silico designed structure consists of a 24-29 bp duplex RNA region linked on both ends by N-alkyl-N dimeric nucleotides (BCn dimers; n = number of carbon atoms of the alkyl chain). A series of N-alkyl-N capped dumbbell-shaped structures were efficiently synthesized by double ligation of BCn-loop hairpins. The resulting BCn-loop dumbbells displayed experimentally higher biostability than their 3′-N-alkyl-N linear version, and were active against a range of mRNA targets. We studied first the effect of the alkyl chain and stem lengths on RNAi activity in a screen involving two series of dumbbell analogues targeting Renilla and Firefly luciferase genes. The best dumbbell design (containing BC6 loops and 29 bp) was successfully used to silence GRB7 expression in HER2+ breast cancer cells for longer periods of time than natural siRNAs and known biostable dumbbells. This BC6-loop dumbbell-shaped structure displayed greater anti-proliferative activity than natural siRNAs.
PubMed: 26975656. Doi: 10.1093/nar/gkw169.