How do membrane proteins fold?
 Membrane-spanning proteins account for 23% of all human genes [1], and numbers are similar for most other organisms [2]. They serve many essential roles in the cell, including solute transport, signal transduction and energy generation. However, our knowledge on how they achieve their functional structure is still scarce. In fact, the biophysical tools used for characterizing the folding and assembly of transmembrane (TM) proteins are limited in comparison to those available for studying soluble proteins. Many experimental assays designed for the study of protein folding are not straightforwardly applicable to membrane proteins because these proteins require the presence of a lipid bilayer, or at least some membrane-mimicking environment (like detergent micelles) to maintain their native structure. This is in contrast to water soluble proteins, for which a generous set of biophysical tools has in many cases allowed the definition of the molecular mechanisms governing their folding and has permitted, in parallel, a better understanding of their function. Thus, there is increasing interest in achieving a similar level of knowledge about the molecular mechanisms that drive the folding of proteins in the membrane environment, and in particular the rules that explain the stability and assembly of TM segments.
Membrane-spanning proteins account for 23% of all human genes [1], and numbers are similar for most other organisms [2]. They serve many essential roles in the cell, including solute transport, signal transduction and energy generation. However, our knowledge on how they achieve their functional structure is still scarce. In fact, the biophysical tools used for characterizing the folding and assembly of transmembrane (TM) proteins are limited in comparison to those available for studying soluble proteins. Many experimental assays designed for the study of protein folding are not straightforwardly applicable to membrane proteins because these proteins require the presence of a lipid bilayer, or at least some membrane-mimicking environment (like detergent micelles) to maintain their native structure. This is in contrast to water soluble proteins, for which a generous set of biophysical tools has in many cases allowed the definition of the molecular mechanisms governing their folding and has permitted, in parallel, a better understanding of their function. Thus, there is increasing interest in achieving a similar level of knowledge about the molecular mechanisms that drive the folding of proteins in the membrane environment, and in particular the rules that explain the stability and assembly of TM segments.
The free energy of transferring hydrophobic TM segments from an aqueous environment into the lipid bilayer provides most of the thermodynamic stability of membrane proteins. In addition, TM segment hydrophobicity is the main factor for driving membrane partitioning. However, hydrophobicity does not rely only on the amino acid composition of the TM segments, but also on structure formation, which in turn depends upon non-trivial atomic-detail interactions with the polypeptide environment. TM segments fold as either α-helices or β-strands, due to the biophysical constraints imposed by the membrane environment [3]. Nevertheless, in biological membranes α-helical membrane proteins are most abundant, and thus they will be the focus of the current text.
The membrane milieu
The chemical and physical properties of the lipid bilayer make it clear that biological membranes provide a very special milieu for proteins. The basic unit of these membranes are lipids, organized in two monolayers with their polar headgroups exposed on the two surfaces and their acyl chains forming a central hydrophobic core. Then, biological membranes are highly heterogeneous along the normal direction, with a large gradient of environmental polarity over a short distance because of steep changes in chemical composition [4]. Additionally, natural membranes have usually a diverse mixture of lipids with different properties, which are asymmetrically distributed between the two bilayer leaflets. The hydrophobicity and thickness of the hydrocarbon core of the membrane bilayer leads to the expectation that membrane-spanning segments minimize the cost of harboring a polar polypeptide backbone by engaging their polar carbonyl and amide groups into a regular pattern of hydrogen bonds. In fact, in order to integrate into the membrane milieu the TM regions adopt extensive secondary structure, most often α-helical conformation.
The road to the membrane

Figure 1. Targeting of membrane proteins to the translocon. A ribosome (yellow) translating the mRNA of a membrane protein is targeted to the membrane through the SRP (purple). SRP recognizes the emerging hydrophobic sequence (red helix), binds to the ribosome and arrests nascent polypeptide elongation. The ribosome/nascent polypeptide/SRP complex binds to the membrane resident SRP receptor (SR, brown), which is associated to the translocon (blue). SRP dissociation from the SR causes the transfer of the hydrophobic sequence to the translocon and the elongation of the nascent polypeptide resumes.
As they do for all other proteins in living organisms, ribosomes translate membrane proteins from their encoding mRNAs. However, the high hydrophobicity of the TM segments present in membrane proteins prevents their synthesis by soluble ribosomes. Instead, the vast majority of membrane proteins are synthesized by membrane-bound ribosomes. When translating an mRNA encoding a membrane protein, the ribosome early synthesizes an N-terminal hydrophobic stretch of amino acid residues (either a true signal sequence or a non-cleavable TM segment) that must navigate through the ribosomal tunnel toward the exit site (Fig. 1). As the nascent polypeptide grows, the signal sequence emerges from the ribosomal exit tunnel and, if it is sufficiently hydrophobic, is recognized by the signal recognition particle (SRP). The SRP binds to the ribosome-nascent polypeptide chain complex, accommodating the signal sequence in an α-helical conformation and slowing down or halting translation (Fig. 1).
Presumably, this gives the SRP some time to find and dock to its partner, the SRP receptor (SR), which is associated to the translocon (a protein-conducting channel). At the docking site, the SR interacts with both the ribosome and the SRP, leading to conformational changes in the SRP that allow the transfer of the ribosome-nascent chain to the translocon [5]. Subsequently, SRP disassembly leads to resumption of translation and, once in the translocon, the nascent chain will deal with membrane insertion.
Protein synthesis by a membrane-bound ribosome

Figure 2. Folding of TM helices inside the ribosome exit tunnel. (A) During the translation of a membrane protein, the physical distance between the P-site of the ribosome and the active site of the ER oligosaccharyl transferase (OST), located nearby the lumenal end of the translocon central pore, sets a minimum distance (d, in number of residues) for nascent polypeptide chain efficient glycosylation. Such a sequence length can be investigated in a glycosylation mapping assays using test sequences in the framework of the model membrane protein Lep from E. coli (see ref. [20] for details about this type of experiment). (B) SDS-PAGE of in vitro translated samples using test sequences of different length (d, values indicated on the top). The test cases shown are based on two native helix fragments of similar length: one hydrophobic (the TM segment of the membrane protein gp41, left) and one hydrophilic (from N-acetylglutamate kinase NAGK, right). The results show that the minimal sequence length for efficient glycosylation (≥50%, i.e., upper bands in the gel with at least equal intensity compared to the lower bands) is larger for the case of the TM segment (at least 71 residues) than for the polar segment (67 residues), indicating that the first one is folded as a helix within the ribosome tunnel while the second adopts likely an extended conformation. (C) Models of characteristic structures obtained upon MD simulations of complexes of the ribosome (in gray) with a nascent chain harboring the TM gp41 sequence (left, green color) or the NAGK sequence (right, orange color). For a complete study with more cases of TM and polar fragments, please see ref. [10].
A ribosome bound to the Endoplasmic reticulum (ER) membrane is more than a mere decoding and synthesizing machine. It is endowed with an exit tunnel through which a newborn membrane protein, constantly growing, navigates toward the translocon to eventually reach its final destination within the bilayer. This molecular corridor creates a specialized microenvironment that allows the ribosome to distinguish TM from secretory segments and direct TM segment integration into the bilayer [6]. One of the features that can modulate the ribosome triage between TM and non-TM segments might be the folding of tethered nascent chains. In fact, folding of TM segments into an α-helical conformation inside the ribosomal exit tunnel has been demonstrated [7–9].
Recently, using in vitro translation of truncated nascent chains trapped within the ribosome tunnel and molecular dynamics simulations, it has been shown that folding within the ribosome is attained for TM, but not for soluble (polar) helices (Fig. 2). The overall hydrophobicity, helicity and length of a given segment have been found to be the major determinants for the identification of TM segments and their eventual adoption of an α-helical structure within the ribosomal exit tunnel [10]. Thus, the ribosome recognizes the TM regions and facilitates a proper environment for their folding, acting in a chaperone-like manner. From the biophysical point of view, preadoption of α-helical conformation could facilitate membrane integration of TM segments upon entering the translocon, which is well positioned below the ribosome to receive the exiting polypeptide chain (Fig. 3).
Membrane insertion
Once within the translocon channel, TM helices have to be transferred laterally to the surrounding lipid bilayer. Insights into the mechanism of membrane insertion have come from both structural studies [11, 12] and molecular dynamics simulations [13]. It is generally accepted that hydrophobicity is the overriding characteristic of TM segments recognized by the translocon to trigger nascent chain insertion [14]. The central component of the translocon, Sec61α in eukaryotes and SecY in prokaryotes and archaea, is itself a membrane protein formed by 10 TM helices arranged around a central pore with a lateral gate for membrane access of polypeptide nascent chains. Upon ribosome binding, lateral gate contacts are weakened and, if the nascent polypeptide sequence allocated in the central pore is sufficiently hydrophobic, the translocon opens laterally allowing access to the lipid bilayer [15].
In this scenario, integration of the first TM segment of a membrane protein into the ER membrane in the correct orientation is considered important in defining the overall topology of an integral membrane protein (Fig. 3). However, the sequential insertion into the membrane of TM segments (one after another) for multi-spanning membrane proteins does not explain the insertion mechanism of all membrane proteins. For instance, it has been demonstrated that poorly hydrophobic sequences insert into the lipid bilayer in a concerted manner as helical hairpins or bundles, as recently reviewed elsewhere [16].
Membrane protein assembly
Once TM helices are established and inserted across the lipid bilayer they interact to form functional tertiary structures (in the case of multi-spanning membrane proteins) (Fig. 3), and in some cases quaternary membrane-spanning structures (not shown). The clues of such complex TM protein-protein interactions are crucial for understanding the biogenesis of membrane proteins, that has been historically neglected due to the difficulties in studying this process experimentally.
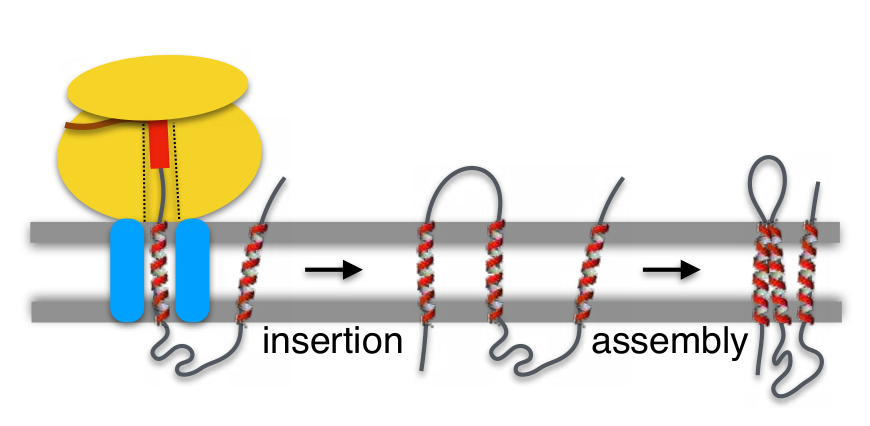
Figure 3. Insertion and assembly of membrane proteins into the membrane. The Insertion of TM segments (red helices) facilitates membrane integration of the newly synthesized protein, which has to be assembled in its native conformation. Monomeric membrane proteins can subsequently associate to form homo- or heteromeric complexes (not shown) to allow the broad variety of key membrane protein activities.
The forces behind TM helix-helix packing are essentially the same as those driving helix packing interactions in soluble proteins. However, their contribution to the folding/packing of the protein is significantly different due to the modified environment (aqueous vs lipidic). In soluble proteins, tertiary and quaternary foldings are mainly driven by the hydrophobic effect and electrostatic interactions. In contrast, in membrane proteins van der Waals interactions have been identified as the primary force behind helix-helix packing. By their nature, van der Waals forces require a large contact area between the associating protein segments. Interestingly, in helical TM segments amino acids with small side chains (like Gly or Ala) are frequently found in helix-helix contact interfaces, while bulky non-polar side chains are located mostly on lipid exposed surfaces. The role of Gly in helix-helix association has been vastly documented in the context of Glycophorin A (GpA), both in membrane-like environments [17, 18] and in cells [19]. The abundance of small residues rather than larger hydrophobic side chains in TM interactions likely reflects the bilayer effect [3] in membranes and the minimal entropic requirements for packing small side chains with few rotatable bonds.
Finally, it remains to be determined whether specific chaperones and/or translocon accessory components facilitate insertion of poorly hydrophobic TM sequences and/or large topological rearrangements needed for the assembly of particular membrane proteins. Considerably more effort will need to be invested in studying the processes underlying membrane protein folding, both in vitro and in vivo, but the way towards our complete understanding starts to be paved.
Membrane Proteins Group, ERI BioTecMed and Depto. de Bioquímica y Biol. Molecular,
Universitat de València, Valencia (Spain).
E-mail: Ismael.Mingarro@uv.es
References
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, et al. “Tissue-based map of the human proteome.” Science, 2015, 347: 1260419. DOI.
- Martínez-Gil L, Saurí A, Marti-Renom MA, Mingarro I. “Membrane protein integration into the endoplasmic reticulum.” FEBS J, 2011, 278: 3846. DOI.
- White SH, Wimley WC. “Membrane protein folding and stability: Physical Principles.” Annu Rev Biophys Biomol Struct, 1999, 28: 319. DOI.
- White SH, von Heijne G. “How Translocons Select Transmembrane Helices.” Annu Rev Biophys, 2008, 37: 23. DOI.
- Jomaa A, Fu Y-HH, Boehringer D, Leibundgut M, Shan S-o, Ban N. “Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.” Nat Commun, 2017, 8: 15470. DOI.
- Lin P-J, Jongsma CG, Liao S, Johnson AE. “Transmembrane segments of nascent polytopic membrane proteins control cytosol/ER targeting during membrane integration.” J Cell Biol, 2011, 195: 41. DOI.
- Woolhead CA, McCormick PJ, Johnson AE. “Nascent Membrane and Secretory Proteins Differ in FRET-Detected Folding Far inside the Ribosome and in Their Exposure to Ribosomal Proteins.” Cell, 2004, 116: 725. DOI.
- Lu J, Deutsch C. “Secondary Structure Formation of a Transmembrane Segment in Kv Channels.” Biochemistry, 2005, 44: 8230. DOI.
- Mingarro I, Nilsson I, Whitley P, von Heijne G. “Different conformations of nascent polypeptides during translocation across the ER membrane.” BMC Cell Biol, 2000, 1: 3. DOI.
- Bañó-Polo M, Baeza-Delgado C, Tamborero S, Hazel A, Grau B, Nilsson I, Whitley P, Gumbart JC, von Heijne G, Mingarro I. “Transmembrane but not soluble helices fold inside the ribosome tunnel.” Nat Commun, 2018, 9: 5246. DOI.
- van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. “X-ray structure of a protein-conducting channel.” Nature, 2003, 427: 36. DOI.
- Egea PF, Stroud RM. “Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes.” Proc Natl Acad Sci USA, 2010, 107: 17182. DOI.
- Gumbart J, Schulten K. “Molecular Dynamics Studies of the Archaeal Translocon.” Biophys J, 2006, 90: 2356. DOI.
- Baeza-Delgado C, von Heijne G, Marti-Renom MA, Mingarro I. “Biological insertion of computationally designed short transmembrane segments.” Sci Rep, 2016, 6: 23397. DOI.
- Voorhees RM, Hegde RS. “Structure of the Sec61 channel opened by a signal sequence.” Science, 2015, 351: 88. DOI.
- Whitley P, Mingarro I. “Stitching proteins into membranes, not sew simple.” Biol Chem, 2014, 395: 1417. DOI.
- Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. “Sequence specificity in the dimerization of transmembrane α-helixes.” Biochemistry, 1992, 31: 12719. DOI.
- Orzáez M, Lukovic D, Abad C, Pérez-Payá E, Mingarro I. “Influence of hydrophobic matching on association of model transmembrane fragments containing a minimised glycophorin A dimerisation motif.” FEBS Lett, 2005, 579: 1633. DOI.
- Grau B, Javanainen M, García-Murria MJ, Kulig W, Vattulainen I, Mingarro I, Martínez-Gil L. “The role of hydrophobic matching on transmembrane helix packing in cells.” Cell Stress, 2017, 1: 90. DOI.
- Tamborero S, Vilar M, Martínez-Gil L, Johnson AE, Mingarro I. “Membrane Insertion and Topology of the Translocating Chain-Associating Membrane Protein (TRAM).” J Mol Biol, 2011, 406: 571. DOI.
