High-Speed Atomic Force Microscopy
Probing the dynamics of biomolecules
 Atomic Force Microscopy (AFM)-based studies constitute today a fairly established methodology to observe the structure of biomolecules and to measure their mechanical properties. However, biomolecules are dynamic in nature; hence, to understand how they work we need to increase the spatiotemporal resolution of conventional AFM. In the last decade, High-Speed AFM (HS-AFM) was developed and successfully applied to several cellular machineries, either cytoplasmic or bound to membranes. The molecular movies obtained by this method provide insights otherwise not accessible by other means to date. The technique itself, alone or combined with other techniques, is in continuous development and its relevance is foreseen to expand in the near future.
Atomic Force Microscopy (AFM)-based studies constitute today a fairly established methodology to observe the structure of biomolecules and to measure their mechanical properties. However, biomolecules are dynamic in nature; hence, to understand how they work we need to increase the spatiotemporal resolution of conventional AFM. In the last decade, High-Speed AFM (HS-AFM) was developed and successfully applied to several cellular machineries, either cytoplasmic or bound to membranes. The molecular movies obtained by this method provide insights otherwise not accessible by other means to date. The technique itself, alone or combined with other techniques, is in continuous development and its relevance is foreseen to expand in the near future.
How it developed
In 2010, the team of Toshio Ando in Kanazawa University (Japan), filmed individual myosin molecules walking on an actin filament [1], operating their AFM instrument at a speed about 1000 times faster than the conventional instruments of that time (Video 1). Besides the visual impact and scientific value of those movies, these experiments illustrated that HS-AFM could obtain concomitantly structural and dynamic data, providing insights unmatched by any other method. But to understand how the development of HS-AFM was reached, let’s walk first briefly through the history of the Scanning Probe Microscopy family.
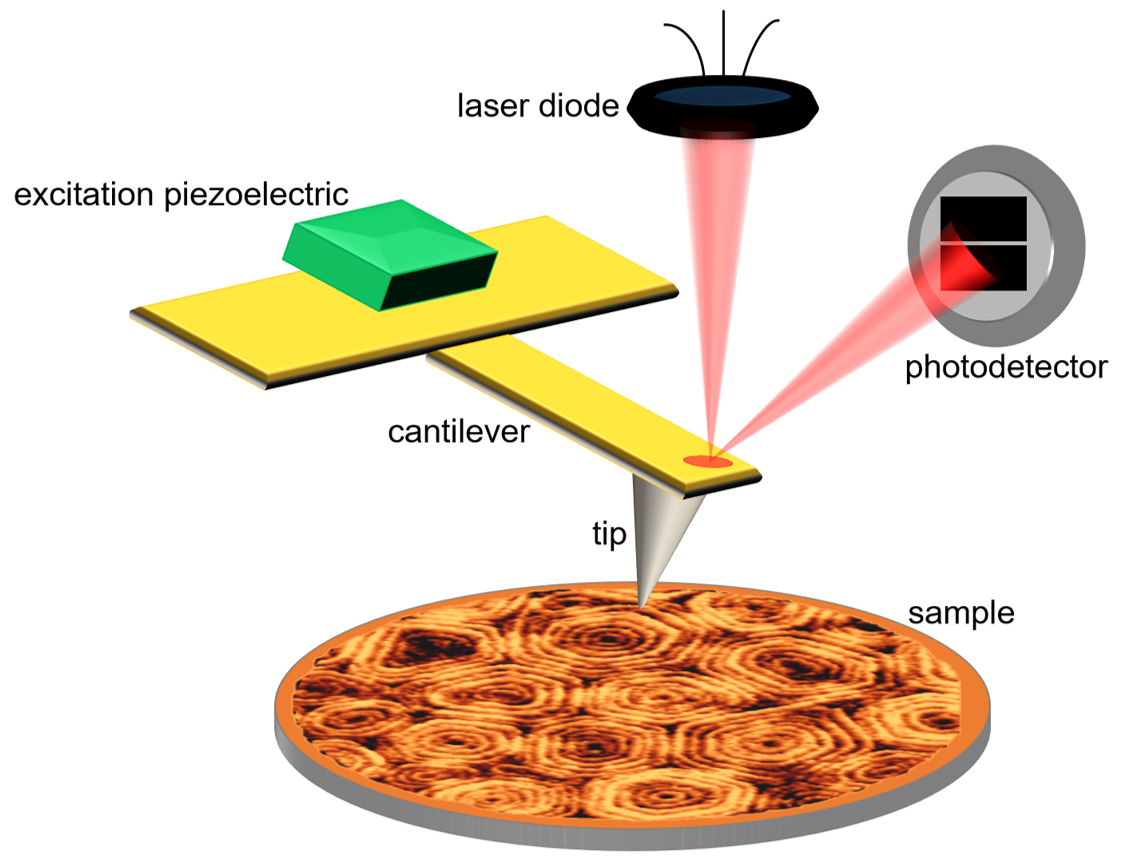
Figure 1. Schematic representation of an AFM instrument operated in tapping-mode. Thanks to a piezoactuator, the AFM cantilever is excited at its resonance frequency. The deflection of the oscillating cantilever is measured by a photodetector which collects the reflection of a laser beam focused at the back of the AFM cantilever.
Scanning Tunneling Microscopy – STM was invented in 1981 [2]. It was a conceptual revolution, because it bypassed the diffraction limit and exhibited a stunning atomic resolution –with higher sensitivity and lower energy consumption than the electron microscope. Five years later, Gerd Binnig and Heinrich Rohrer were awarded the Nobel Prize in Physics for the invention of the STM. As STM measures the tunneling current between a metallic tip and the sample surface, it requires this surface to be conductive. Therefore, it was not particularly suitable for biological samples –despite some circumvents, like covering the sample with a carbon or metal film. By 1986, the requirement about conductivity was overcome when Gerd Binning, Calvin Quate and Cristopher Gerber developed a variation of the STM: Atomic Force Microscopy, AFM [3]. The AFM tip, which resembles a nanometer-sharp needle of a “record player” attached to a (micro)cantilever, feels the sample surface while scanning it using piezoelectric elements. An optical system is used to monitor the cantilever deflection (or force), thanks to a laser beam reflected on the back of the cantilever and detected by a photodiode. The change in deflection due to surface protrusions while the tip scans the sample is translated into electrical signals, thanks to a feedback mechanism. The AFM provides submolecular resolution of the topography of any type of surface, and can operate in aqueous environment on a wide variety of biological samples, from single molecules, such as DNA or proteins, to macromolecular assemblies, cells or tissues.
And yet, it moves
The frame rate of conventional AFM of one image every few minutes is not sufficient to visualize most dynamic processes of biological molecules, which take place in the majority of cases at sub-second time scales. Unlike other structural techniques that rely on ensemble averaging, such as X-ray crystallography, NMR or electron microscopy, single-molecule optical microscopy techniques are particularly well suited for the study of the dynamics of molecules. However, these latter methods rely on detection of fluorescent markers attached to proteins or lipids, which means that the resolution is limited to ~200 nm due to diffraction of light and to a few tens of nm thanks to super-resolution techniques. Besides, some other current techniques are pushing the limits in temporal resolution. This is the case, for example, of time-resolved serial femtosecond crystallography with X-ray free electron laser [4], although it is only pertinent to certain types of samples.The frame rate limitation of conventional AFM was only practical
In this scenario, AFM was a good candidate to occupy a niche in the myriad of biophysical techniques that tackle the dynamic visualization of molecules. The frame rate limitation of conventional AFM was not theoretical, but only practical. In the 1990s, two labs independently started efforts to achieve high speed AFM: Paul Hansma’s group [5] in Santa Barbara (CA, USA) and Toshio Ando’s group. The latter obtained a prototype in 2001 [6] which was practically developed for routine studies of biological samples in 2008 [7]. Currently, its functionality is being expanded, by applying different imaging modes, but mostly in combination with other techniques, such as fluorescence microscopy [8].
Optimization of the imaging rate
HS-AFM has successfully filmed a wide variety of samples, including proteins in action, membranes and cellsFor the study of biological samples, it is common to use AFM modes that have an intermittent tip-sample contact. Among them, the most often used is tapping-mode, in which the AFM cantilever is excited at its resonance frequency. The resulting oscillating tip is intermittently contacting -tapping- the surface, giving rise to a damped oscillation amplitude. The topography of the surface is reconstructed by monitoring the amplitude of the oscillating cantilever (Fig. 1). While scanning in the X-Y direction, the amplitude of the cantilever oscillation is kept constant thanks to a feedback control. HS-AFM is nowadays almost exclusively operated in tapping-mode to minimize the force or amount of energy caused by the tip-sample contact, at maximum rates of 5-10 frames per second (fps) [9]. See for example the rotation and diffusion of the outer membrane protein F (OmpF) inserted in a lipid membrane (Video 2). In the video, acquired at ~2 fps, one can follow the organization of the trimeric structure of these channels in near-native conditions [10]. The technical developments which allow high-speed rates are based on the miniaturization of the moving components of the AFM (cantilever and scanner) to increase their velocity by 1000 times and thus be able to achieve reaction responses of microseconds. Below, I briefly detail the most important elements that allow the AFM to be operated at a video-rate:
- Small cantilevers: The cantilever dimensions were reduced to 7 µm long, 2 µm wide and 90 nm thick (with respect to conventional cantilevers, 60-200 µm long, 100-170 µm wide and 400-800 nm thick). This point is crucial: the reduction in mass of these small microfabricated cantilevers allows for higher resonance frequencies and shorter response times (~1 µs). In order to get high-resolution imaging, long amorphous carbon deposits are grown on the cantilever, which serve as a tip once etched.
- Fast scanners: The z-piezo actuator is subjected to high-frequency displacements during high-speed imaging, hence it tends to generate mechanical vibrations. Thus, a dummy piezo is placed in the opposite direction and displaced simultaneously with the same length in order to counterbalance the impulsive force and therefore minimize the vibrations. Furthermore, each z-scanner is calibrated and the spurious frequencies are filtered out. The x- and y- piezo actuators are also smaller than in conventional AFM. Their center of masses are kept stationary using flexures to hold them and by attaching a balance weight to the counter side. They are also embedded in a silicon elastomer to passively damp vibrations.
- Fast amplitude detectors: The cantilever oscillation amplitude is measured and output by a Fourier method at every cycle of the oscillation.
- Adaptive feedback: During fast imaging, the tip tends to “detach” completely from the surface at the downhill regions, an effect known as parachuting. To address this, a dynamic controller was developed, consisting in a Proportional-Integral-Derivative (PID) feedback controller that increases automatically the gain when downhill regions are scanned.
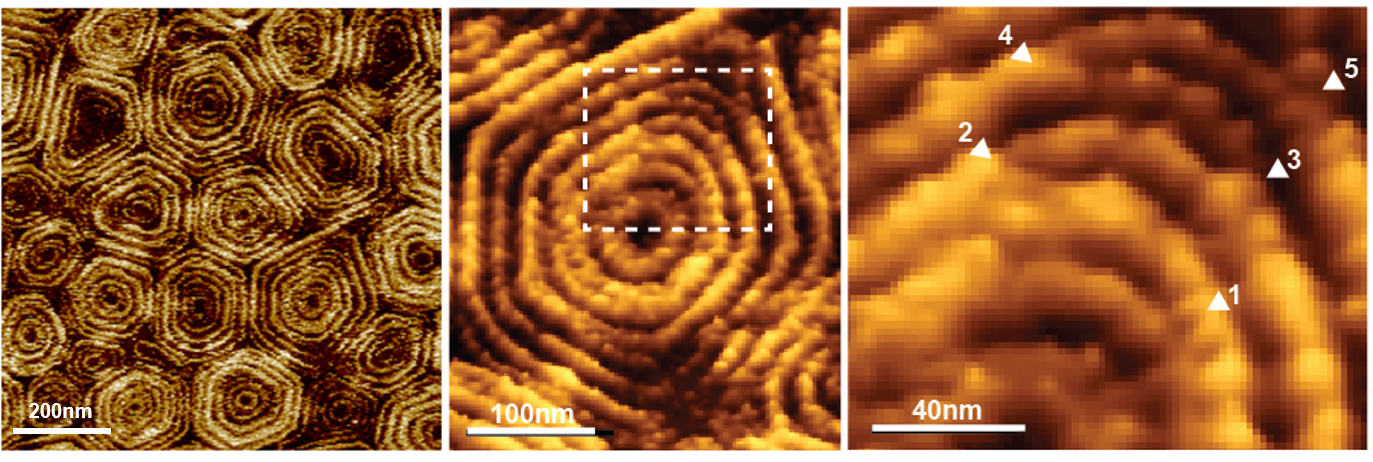
Figure 2. High-resolution AFM topographic image of a membrane area containing Snf7 patches. A large number of patches are shown in the panel on the left and one of them is enlarged in the center panel. The square drawn in this enlargement was zoomed and is shown on the right, where it reveals splits and variability of Snf7 structures. Reproduced from: Chiaruttini et al. Cell 2015, 163: 866 [13], CC BY NC ND 2015.
Still, we have to keep in mind that AFM is a surface technique, so the molecules of interest need to be adsorbed or be immobilized in a substrate. This, in turn, may restrict the natural interaction between molecules. Thus, for each system to be studied by HS-AFM, the conditions of sample, medium and substrate need to be optimized. Even so, molecules diffusing very fast on the surface can not be imaged by means of HS-AFM. Nevertheless, HS-AFM has successfully filmed a wide variety of samples, mostly proteins in action, but also membranes and cells, which has provided important insights about these systems. To review recent reports of dynamic imaging by HS-AFM see [11, 12].
Sample case: HS-AFM imaging of membrane remodeling processes
HS-AFM has been recently fruitful to study membrane remodeling processes, particularly fission machineries. We studied the ESCRT-III (Endosomal Sorting Complex Required for Transport) system [13]. ESCRT-III is needed for lipid membrane remodeling in many cellular processes, from abscission to viral budding and the formation of late endosomes. However, how ESCRT-III polymerization generates membrane curvature remains debated. We showed that Snf7, the main component of ESCRT-III, polymerized into spirals at the surface of supported lipid bilayers (Fig. 2).
The spring-like activity of ESCRT-III could be the driving force for mediate membrane remodelingWe were able to record HS-AFM movies of the Snf7 complex formation and its dynamics from filament to the maturated circular assembly around the membrane constriction site. We observed interfilament dynamics that provides a basis for a mechanistic explanation of how this protein machinery creates tension for membrane fission (Video 3). Furthermore, we showed that Snf7 assemblies compress the inner diameter during maturation, which constitutes a direct evidence of force generation during the assembly process. When we disrupted the spirals with the AFM cantilever, the broken polymers spontaneously rearranged forming smaller rings, suggesting a preferred curvature of the Snd7 polymer of ~25 nm. We reasoned that Snf7 spirals could function as spiral springs. From measurements of the polymerization energy and the rigidity of Snf7 filaments, we concluded that they were deformed while growing in a confined area.
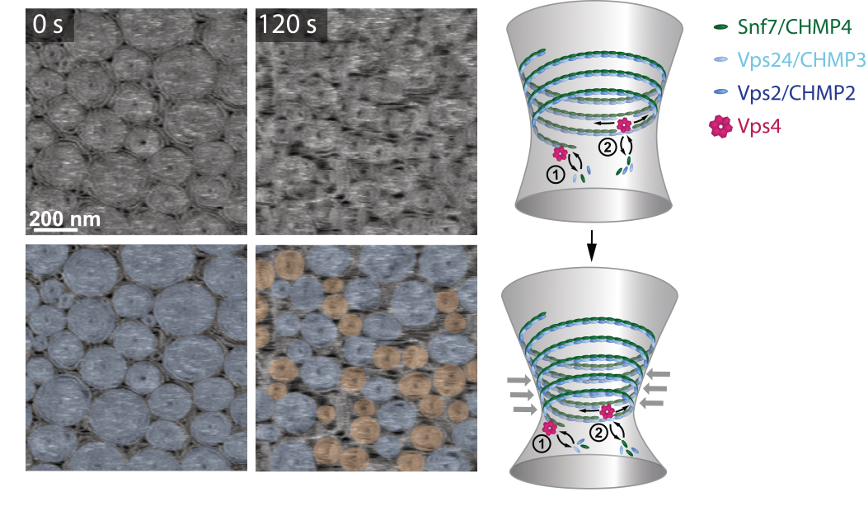
Figure 3. Vps4 induces a dynamic reorganization of ESCRT-III assemblies. HS-AFM imaging of ESCRT-III polymers on supported lipid membranes, followed by the addition of Vps4 and ATP. The overlays highlight pre-formed spirals (blue) or newly formed spirals (orange). Our model suggests that Vps4 mediates continuous subunit turnover in ESCRT-III assemblies during growth and constriction. Reproduced by permission from Springer Nature: Mierzwa et al. Nat Cell Biol 2017, 19: 787 [15], Copyright 2017.
Furthermore, our dynamic data also suggest that the elastic expansion of compressed Snf7 spirals could stretch the lipids they are bound to, generating an area difference between the membrane leaflets and thus inducing curvature [13, 14]. This spring-like activity can be the driving force by which ESCRT-III could mediate membrane remodeling, which is a new property in this field.
In our most recent work, we studied the molecular role of Vps4, an ATPase that it is known to drive the disassembly of persisting filaments of ESCRT-III [15]. We observed that in the presence of Vps4, ESCRT-III polymers disassemble partially, remaining the most-inner part of the ring-like structure refractory to the action of the enzyme. Surprisingly, in the presence of a soluble Snf7 pool, ESCRT-III assemblies shrink under the action of Vps4, liberating free space in the membrane were new ESCRT-III assemblies are growing simultaneously (Fig. 3 and Video 4). This results in a high exchange and lateral mobility of ESCRT-III assemblies on membranes. The dynamic exchange provides an explanation for how ESCRT-III filaments gradually adapt their shape during membrane constriction, which has broad implications in diverse cellular processes, differing in size, shape and duration –such as plasma membrane repair, cytokinesis or viral budding.
Beyond imaging: from molecular to cellular mechanics

Figure 4. HS-FS of titin unfolding. (a) Schematic process of titin forced unfolding showing the relevant steps: (1) relaxed polyprotein, (2) polyprotein stretching, (3) unfolding of one domain and (4) unfolded domain stretching. Two examples of force extension curves revealing three unfolding peaks at 1 μm/s (bottom) and 1000 μm/s (top) are also shown. (b) Most probable force versus retraction velocity of titin unfolding using HS-FS (●), conventional AFM (□) and MD simulations (△). The red line is the fit of a model describing the response of the unfolding energy landscape. Reproduced by permission from John Wiley and Sons: Eghiaian et al. FEBS Lett 2014, 588: 3631 [12], Copyright 2014.
The flexibility of the AFM cantilever also allows applying picoNewton forces to the sample. When the AFM is operated in force spectroscopy mode, the tip approaches and retracts from the surface with controlled force and velocity. This operation mode allows probing the mechanical properties of single molecules, membranes and living cells. Félix Rico et al. adapted the HS-AFM system to perform force spectroscopy measurements on single titin domains, which was unfolded at high velocities with microsecond time resolution [16]. Remarkably, the speed of these HS-AFM measurements -up to ~4 mm/s- reached that of molecular dynamics simulations, bridging the gap between the experiment and computational approaches and providing further insights into the mechanical behavior of biomolecules (Fig. 4). Most recently, the HS-AFM system was further adapted to study the microrheology of living cells over a wide dynamic range of frequencies [17].
Conclusions: progress and perspectives of the HS-AFM technique
The possibilities of HS-AFM are still emerging and expanding. Some of the latest efforts are dedicated to observe dynamic and morphological changes in live cells, which imply the development of fast-wide area scanners and very long electron-beam deposited tips [18]. However, much of the future of HS-AFM seems to rely on its coupling with other techniques. The company RIBM in Japan has developed a tip-scan (not sample-stage scan) HS-AFM in order to correlate it with light microscopy. Fukuda et al. already have coupled it to a total internal reflection microscope [19] but the combination to other super-resolution optical microscopies remains to be explored. The team of Toshio Ando is working to combine HS-AFM with optical tweezers to visualize proteins under the application of an external force. Other advances will focus on the optimization of HS-AFM for different imaging modes; for example, to image in so-called multimodal (exciting the cantilever at two or more resonance frequencies) or realize a high-speed mechanical mapping (a force curve in every pixel of the image). Such current and future developments could have a broad impact in biophysics, as they will likely allow further exploring the mechano-transduction pathways in cells and help opening new fields of discovery in basic and applied biological research.
N.B.: I want to finish by noting that a significant amount of researchers pushing the HS-AFM methodology in European biophysics were, like myself (currently in Lille, France), formed in Spain. This is the case of my colleagues Félix Rico and Ignacio Casuso (in Marseille, France), Adai Colom (Aurélien Roux’s lab in Geneva, Switzerland), Laura Picas (Montpellier, France) and Ignacio Munguira (Wouter Roos’ team in Groningen, Netherlands).
Lorena Redondo-Morata
Force Microscopy for Biophysics Lab,
Institut de la Santé et la Recherche Médicale – Inserm U1006,
Aix-Marseille Université – AMU, Marseille (France).
Current address:
Cellular Microbiology and Physics of Infection Lab,
Institut de la Santé et la Recherche Médicale – Inserm U1019,
Institut Pasteur Lille, Marseille (France).
E-mail: lorena.redondo@inserm.fr
Movies
Video 1. Processive movement of myosin V under ATP hydrolysis, captured at 7 fps. Scan range, 130 nm × 65 nm. Reproduced by permission from Annual Reviews: Ando, Uchihashi & Kodera. Annu Rev Biophys 2013, 42: 393 [20], Standard YouTube License 2013.
Video 2. Diffusion of the bacterial outer membrane protein F (OmpF) on the membrane. The image size is 7.5 nm and was acquired at ~2 fps. Reproduced by permission from Springer Nature: Casuso et al. Nat Nanotechnol 2012, 7: 525 [10], Copyright 2012. Courtesy of Ignacio Casuso.
Video 3. Birth of new Snf7 spiral assemblies on a lipid bilayer. The frame size is 350 nm and was acquired at 1.2 fps. Reproduced from: Chiaruttini et al. Cell 2015, 163: 866 [13], CC BY NC ND 2015.
Video 4. Vps4 under ATP hydrolysis causes a non-equilibrium growing and shrinking behavior of concentric rings and spirals in 2D. The movie shows a constant turnover of subunits between the solution and the filaments. The frame size is 800 nm and the imaging speed 1 fps. Reproduced by permission from Springer Nature: Mierzwa et al. Nat Cell Biol 2017, 19: 787 [15], Copyright 2017.
References
- Kodera N, Yamamoto D, Ishikawa R, Ando T. “Video imaging of walking myosin V by high-speed atomic force microscopy.” Nature, 2010, 468: 72. DOI.
- Binnig G, Rohrer H, Gerber C, Weibel E. “Surface Studies by Scanning Tunneling Microscopy.” Phys Rev Lett, 1982, 49: 57. DOI.
- Binnig G, Quate CF, Gerber C. “Atomic Force Microscope.” Phys Rev Lett, 1986, 56: 930. DOI.
- Nango E, Royant A, Kubo M, Nakane T, Wickstrand C, Kimura T, Tanaka T, Tono K, Song C, Tanaka R, Arima T, Yamashita A, et al. “A three-dimensional movie of structural changes in bacteriorhodopsin.” Science, 2016, 354: 1552. DOI.
- Viani MB, Pietrasanta LI, Thompson JB, Chand A, Gebeshuber IC, Kindt JH, Richter M, Hansma HG, Hansma PK. “Probing protein–protein interactions in real time.” Nat Struct Biol, 2000, 7: 644. DOI.
- Ando T, Kodera N, Takai E, Maruyama D, Saito K, Toda A. “A high-speed atomic force microscope for studying biological macromolecules.” Proc Natl Acad Sci U S A, 2001, 98: 12468. DOI.
- Ando T, Uchihashi T, Fukuma T. “High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes.” Prog Surf Sci, 2008, 83: 337. DOI.
- Colom A, Casuso I, Rico F, Scheuring S. “A hybrid high-speed atomic force–optical microscope for visualizing single membrane proteins on eukaryotic cells.” Nat Commun, 2013, 4: 2155. DOI.
- Ando T. “High-speed atomic force microscopy coming of age.” Nanotechnology, 2012, 23: 062001. DOI.
- Casuso I, Khao J, Chami M, Paul-Gilloteaux P, Husain M, Duneau J-P, Stahlberg H, Sturgis JN, Scheuring S. “Characterization of the motion of membrane proteins using high-speed atomic force microscopy.” Nat Nanotechnol, 2012, 7: 525. DOI.
- Ando T. “Directly watching biomolecules in action by high-speed atomic force microscopy.” Biophys Rev, 2017, 9: 421. DOI.
- Eghiaian F, Rico F, Colom A, Casuso I, Scheuring S. “High-speed atomic force microscopy: Imaging and force spectroscopy.” FEBS Lett, 2014, 588: 3631. DOI.
- Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, Roux A. “Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation.” Cell, 2015, 163: 866. DOI.
- Roux A. “ESCRT-III hypothetical mechanisms.” YouTube, 00:01:04 (2015). ID: wdT14aQ-Ag4
- Mierzwa BE, Chiaruttini N, Redondo-Morata L, von Filseck JM, König J, Larios J, Poser I, Müller-Reichert T, Scheuring S, Roux A, Gerlich DW. “Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis.” Nat Cell Biol, 2017, 19: 787. DOI.
- Rico F, Gonzalez L, Casuso I, Puig-Vidal M, Scheuring S. “High-Speed Force Spectroscopy Unfolds Titin at the Velocity of Molecular Dynamics Simulations.” Science, 2013, 342: 741. DOI.
- Rigato A, Miyagi A, Scheuring S, Rico F. “High-frequency microrheology reveals cytoskeleton dynamics in living cells.” Nat Phys, 2017, 13: 771. DOI.
- Shibata M, Uchihashi T, Ando T, Yasuda R. “Long-tip high-speed atomic force microscopy for nanometer-scale imaging in live cells.” Sci Rep, 2015, 5: . DOI.
- Fukuda S, Uchihashi T, Iino R, Okazaki Y, Yoshida M, Igarashi K, Ando T. “High-speed atomic force microscope combined with single-molecule fluorescence microscope.” Rev Sci Instrum, 2013, 84: 073706. DOI.
- Ando T, Uchihashi T, Kodera N. “High-Speed AFM and Applications to Biomolecular Systems.” Annu Rev Biophys, 2013, 42: 393. DOI.