Ancestral proteins: How and why
 Strictly speaking, ancestral proteins are proteins from extinct organisms. However, neither extinct organisms nor their proteins exist any more, which raises two questions. First, can we bring ancient proteins back? De-extinction of, for instance, the woolly mammoth or the passenger pigeon, is being discussed as a real possibility for a not too distant future [1]. A protein is certainly a much simpler system than a whole animal. But, and this is the second question, why should we want to bring ancient proteins back to life in any case (besides the fact that some people may think that it is a cool idea)?
Strictly speaking, ancestral proteins are proteins from extinct organisms. However, neither extinct organisms nor their proteins exist any more, which raises two questions. First, can we bring ancient proteins back? De-extinction of, for instance, the woolly mammoth or the passenger pigeon, is being discussed as a real possibility for a not too distant future [1]. A protein is certainly a much simpler system than a whole animal. But, and this is the second question, why should we want to bring ancient proteins back to life in any case (besides the fact that some people may think that it is a cool idea)?
Can we bring ancestral proteins back? Yes, but with some peculiaritiesAs to the first question (can we bring ancestral proteins back?), the answer is certainly yes, but with some peculiarities that must be noted. De-extinction of the woolly mammoth would require, first of all, finding mammoth DNA in preserved tissue remains. This is a real possibility since mammoths became extinct a few thousand years ago. However, with a few exceptions, individual proteins of this age are not expected to be of much interest by themselves because of the small sequence differences with their modern counterparts. Proteins from organisms that existed millions or even billions of years ago, on the other hand, will display substantial sequence differences with modern proteins and are a priori more interesting targets for protein de-extinction. However, finding useful DNA in fossils of those ages is extremely unlikely.
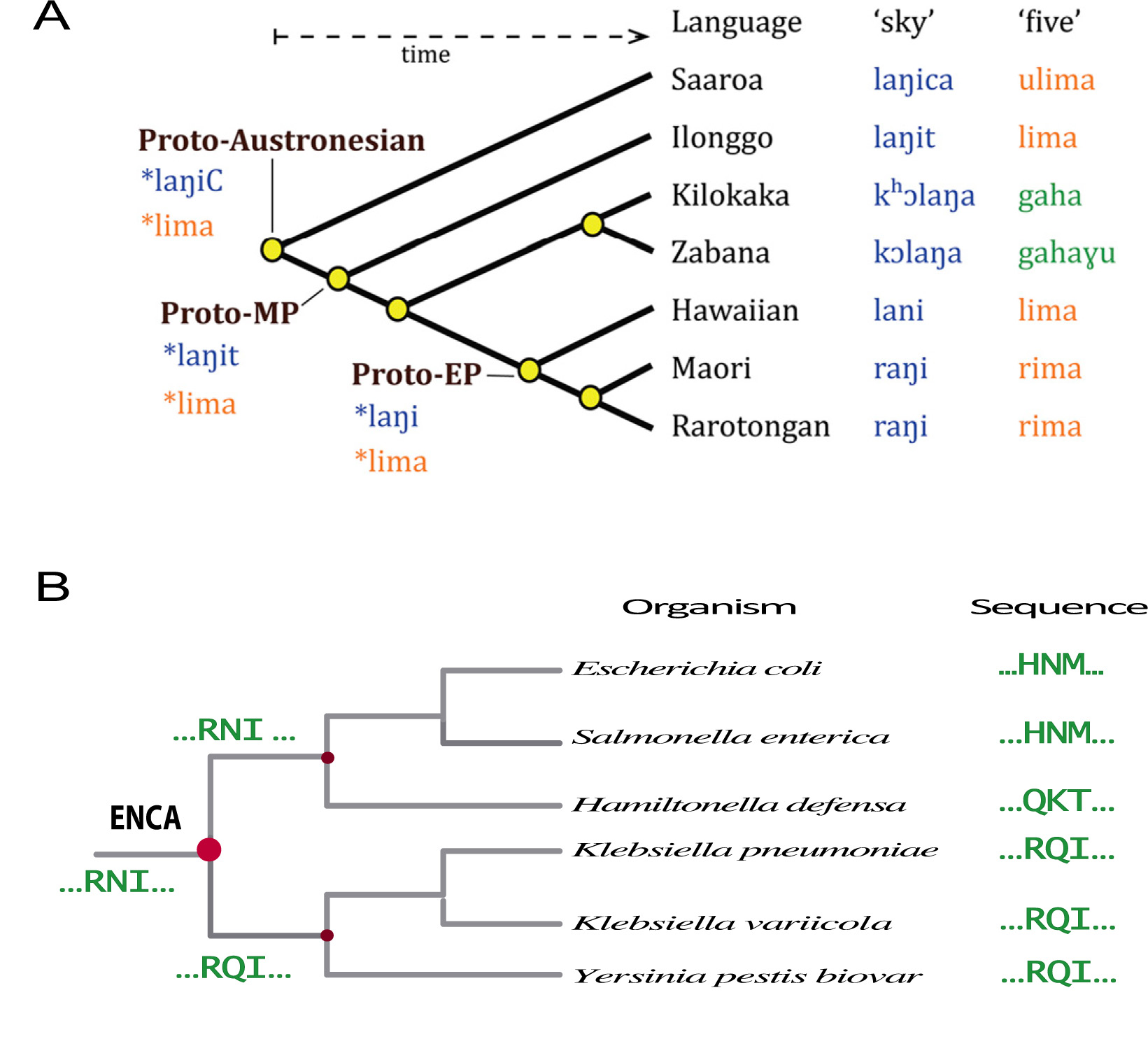
Figure 1. Reconstruction of the sequences of extinct proteins is similar is similar to reconstruction of words from extinct languages. (A) Plausible approximations to words in extinct languages can be derived from the corresponding words in modern, related languages by using suitable models of language evolution. Reprinted, with permission, from Atkinson. Proc Natl Acad Sci USA 2013, 110: 4159 [2]. (B) Protein sequences can be viewed as words written using an alphabet of 20 letters; plausible approximations to ancestral protein sequences can thus be derived from the sequences of their modern descendants by using suitable phylogenetic and bioinformatics analyses. Part of a phylogenetic for β-lactamase evolution is shown for illustration. Only a small section of the protein sequence is shown. ENCA stands for the last common ancestor of Enterobacteria.
On the other hand, phylogenetic and bioinformatics analyses of modern protein sequences can lead to plausible approximations to the sequences of their ancestors. This process is similar to the reconstruction of words of extinct languages (Fig. 1) from the words in modern languages by using suitable models of language evolution [2]. Of course, once ancestral sequences are available, standard molecular biology methodologies can be used to prepare in the laboratory the proteins encoded by the reconstructed sequences. In the jargon of the field, this second step of the process is referred to as ancestral protein resurrection. At the time of writing, over 50 protein systems have been studied using sequence reconstruction followed by laboratory resurrection [3] and some of these studies have targeted phylogenetic nodes close to last common ancestor of life. Admittedly, what these studies have brought back to life are only plausible approximations to the proteins that existed long time ago.
Resurrected ancestral proteins helps addressing problems in evolutionAs to the second question (why should we want to resurrect ancestral proteins?) there are actually two quite different, but also quite convincing, answers. First, research carried out in the last ~25 years has demonstrated that resurrected ancestral proteins may provide useful tools to address important problems in evolution. Secondly, more recent work has emphasized the potential biomedical and biotechnological implications of ancestral protein resurrection. Illustrative examples of these two applications are briefly described below.
Many people consume alcoholic drinks on a regular basis and some of them eventually develop serious health conditions related with alcohol consumption. One possible explanation for our problems with alcohol is simply that alcohol has appeared recently in our diet and that, therefore, we have not had time to adapt to it. One could argue that the incorporation of alcohol in the diet is a consequence of the development of agriculture and the use of fermentation to process food. Agriculture originated a few thousand years ago, which is indeed a very short time in evolutionary terms. Benner and coworkers [4] recently resurrected ancestral alcohol dehydrogenases and found an increase in the ability of these enzymes to degrade alcohol at about 10 million years ago. This result supports a quite different evolutionary narrative. There is evidence that, about 10 million years ago, our ancestors left the top of the trees for the floor of the jungle and, consequently, gained access to fruit with a significant content of alcohol (i.e., fruit dropped from the trees that had undergone fermentation). It is likely, overall, that alcohol appeared in the diet of our ancestors millions of years before the development of agriculture.

Figure 2. Evolutionary history of uricases as revealed by ancestral protein resurrection. Numbers in red are a metric of uricase activity. A gradual decrease in activity is observed before the pseudogenization events that led to the lack of uricase in humans and other primates. The decrease likely allowed our ancestors to accumulate fat from the metabolism of fructose (see text for details). This figure is modified with permission from figures originally published in Kratzer et al. Proc Natl Acad Sci USA 2014, 111: 3763 [5].
In a similar study, Gaucher and coworkers [5] used ancestral protein resurrection to gain insight into the evolutionary origin of high uric acid levels in humans. The immediate cause of hyperuricemia is of course known: we, humans, do not synthesize uricase, the enzyme that degrades uric acid in most animals. This is linked to the pseudogenization of the uricase gene due, among other alterations, to a mutation that introduced a stop codon. This seemingly simple explanation poses, however, additional questions in an evolutionary context. At the molecular level, evolution is, to some substantial extent, purifying natural selection that efficiently eliminates, at least in large populations, deleterious mutations. It is, therefore, puzzling that inactivating alterations of the uricase gene occurred in the line of descent that leads to humans.
One possible solution is that high levels of uric acid are actually advantageous under some circumstances. However, it is not at all clear what these advantages may be. In fact, all the consequences of high uric acid levels in humans (gout, kidney stones, etc.) appear to be harmful. To clarify these issues, Gaucher and coworkers resurrected ancestral uricases and determined their activity levels (Fig. 2). They found that the capability of uricase to degrade uric acid had continuously decreased before the events that led to the pseudogenization of the uricase gene. This decrease occurred at the end of the Oligocene, a period of environment cooling which made it difficult for our ancestors to find fruit, their likely staple food. However, uric acid facilitates the accumulation of fat from the metabolism of fructose by upregulating some of the enzymes involved.

Figure 3. Ancestral enzymes in modern organisms. (A) Schematic phylogenetic tree used in the reconstruction of ancestral thioredoxins. (B) Replacement of E. coli thioredoxin with resurrected ancestral thioredoxin impairs E. coli fitness, as revealed by generation time determinations; a gradual dependence with evolutionary distance is, however, observed and some ancestral thioredoxins show substantial functionality within E. coli. (C) Bacteriophage T7 recruits thioredoxin (red) for its replisome, where it is involved in a strong and specific interaction with the thioredoxin binding domain of the virus polymerase gp5 (green). (D) Most ancestral thioredoxins cannot be recruited and block virus propagation. Figures in panels B and D are from Delgado et al. Cell Rep 2017, 19: 1247 [6], Open Access (CC BY-NC-ND 4.0 2017).
High levels of uric acid helped, therefore, our ancestors survive periods of fruit scarcity. Of course, the conditions that conferred some advantage to high uric acid levels are no longer relevant, at least in developed countries. However, there is no simple way to unevolve the mutational changes that led first to a decrease in uricase activity and then to the pseudogenization of the uricase gene. We are, therefore, stuck with high uric acid levels and its harmful consequences.
A given ancestral protein may preserve normal functionality and, at the same time, prevent infection by a given pathogenProteins often interact in vivo with a large number of macromolecular components. Replacing a modern protein within a modern organism with one of its resurrected ancestors will, therefore, affect many biologically relevant intermolecular interactions. There are, however, two sides to this coin. On the one hand, organismal fitness may be impaired by the replacement because the in vivo functionality of the replaced protein and its interacting partners will be compromised. On the other hand, the replacement might help the organism evade pathogens. The reason is that pathogens and their hosts co-evolve and a successful pathogen has evolved to efficiently recruit the proteins of its host for its own purposes. The possibility arises then that a given ancestral protein may hit the sweet spot where the normal functionality is preserved to a significant extent and, at the same time, a given pathogen is prevented from infecting the organism.

Figure 4. Enhanced stability and substrate promiscuity in resurrected ancestral lactamases. (A) Ancestral and modern lactamases share the canonical lactamase fold. (B) Still, lactamases from 2-3 billion-year phylogenetic nodes show denaturation temperatures 30-35 degrees above the typical values for modern mesophilic lactamases. (C) Furthermore, the “oldest” lactamases can degrade efficiently different lactam antibiotics (BF: benzylpenicillin; CTX: cefotaxime; CAZ: ceftazidime) while the modern TEM-1 lactamase is a penicillin specialist that shows very low activity levels with third-generation antibiotics (such as CTX and CAZ). This promiscuity is linked to conformational diversity/flexibility as shown by NMR-relaxation studies (D), which show a larger number of residues with dynamic contributions in the μs-ms time range (labeled in red), and by molecular dynamics simulations (E). Figures in panels A, B and C are reproduced with permission from Risso et al. J Am Chem Soc 2013, 135: 2899 [7], Copyright 2013, American Chemical Society. Figures in panels D and E are reproduced from Risso et al. Nat Commun 2017, 8: 16113 [9], Open Access (CC BY 4.0 2017).
To explore this possibility, we recently replaced the thioredoxin within E. coli with several resurrected Precambrian thioredoxins [6]. Thioredoxins are general oxido-reductases in all known cells but, in addition, E. coli thioredoxin is a proviral factor for the phage T7, a virus that infects E. coli. The phage recruits thioredoxin for its replisome where it binds strongly and specifically to the viral gp5 polymerase (Fig. 3), an interaction that is essential for replisome efficiency. Some resurrected ancestral thioredoxins showed acceptable levels of functionality within E. coli as revealed by the determination of generation times, but could not be recruited by the virus for its replisome, thus preventing virus propagation within E. coli. More generally, these results suggest an approach to the engineering of pathogen-resistant crops.
Instead of using modern proteins as starting point for engineering, we would use resurrected ancestral proteinsResurrected ancestral proteins may display properties that are useful in scaffolds for protein engineering. Enhanced stability, for instance, is a common outcome of ancestral resurrection, likely linked to the thermophilic nature of primordial life. Also, resurrected ancestral enzymes are often found to be able to catalyze several more or less related reactions. This catalytic promiscuity may reflect the generalist nature of primordial enzymes or may be the result of having targeted pre-duplication nodes in the evolution of new functions. In any case, promiscuity is likely linked to conformational flexibility/diversity, i.e., to the capability to populate a diversity of conformations and it is a feature that, together with enhanced stability, should contribute to protein evolvability. That is, enhanced stability and conformational diversity contribute to the capability of a protein scaffold to generate new functionalities by allowing functionally useful but destabilizing mutations to be accepted and by promoting the efficient search of functionally competent conformations [7, 8].

Figure 5. Using resurrected ancestral lactamases as scaffolds for the engineering of new active sites. (A) Schematic representation of the phylogenetic tree used for the reconstruction of ancestral lactamase sequences. (B) A simple minimalist design based on 1-2 mutations generates de novo catalysis for the Kemp elimination when using the ancestral lactamases as scaffolds; the figure shows the new active site generated with a bound transition-state analogue. (C) The levels of catalysis achieved compare well with previous rational designs and are less than two orders of magnitude from the best Kemp eliminase reported to date which was the outcome of 17 rounds of directed evolution. Reproduced from Risso et al. Nat Commun 2017, 8: 16113 [9], Open Access (CC BY 4.0 2017).
A few years ago [7], we found resurrected ancestral lactamases to combine these two features (Fig. 4) and, more recently [9], we have used a simple minimalist design to generate a de novo enzyme functionality in these ancestral lactamases (Fig. 5). Remarkably, the same minimalist approach consistently failed when we used modern lactamases as scaffolds for engineering. The engineering of new enzymes capable of catalyzing non-natural reactions is one of the most important unsolved problems in protein science. It is also a goal with enormous biotechnological implications. Our results [9] suggest that a molecular version of Gould’s “replaying the tape of life” experiment [10] could provide a useful approach in this context. That is, instead of using modern proteins as starting point for engineering, we would use resurrected ancestral proteins. Modern proteins are often highly specialized and it is difficult to teach an old dog new tricks. On the other hand, some resurrected ancestral proteins at least represent early stages of molecular evolution at which new functionalities were generated. Plausibly, their evolution could then be replayed in the laboratory and directed towards new functionalities of biotechnological interest. This is indeed an exciting possibility that it is being actively pursued by several groups in the field.
Departamento de Química Física,
Universidad de Granada,
Granada (Spain).
E-mail: sanchezr@ugr.es
Departamento de Química Física,
Universidad de Granada,
Granada (Spain).
E-mail: vrisso@ugr.es
References
- Wray B, Rise of the necrofauna. The science, ethics and risks of de-extinction. Greystone Books, 2017. URL.
- Atkinson QD. “The descent of words.” Proc Natl Acad Sci USA, 2013, 110: 4159. DOI.
- Gumulya Y, Gillam EMJ. “Exploring the past and the future of protein evolution with ancestral sequence reconstruction: the ‘retro approach to protein engineering.” Biochem J, 2016, 474: 1. DOI.
- Carrigan MA, Uryasev O, Frye CB, Eckman BL, Myers CR, Hurley TD, Benner SA. “Hominids adapted to metabolize ethanol long before human-directed fermentation.” Proc Natl Acad Sci USA, 2014, 112: 458. DOI.
- Kratzer JT, Lanaspa MA, Murphy MN, Cicerchi C, Graves CL, Tipton PA, Ortlund EA, Johnson RJ, Gaucher EA. “Evolutionary history and metabolic insights of ancient mammalian uricases.” Proc Natl Acad Sci USA, 2014, 111: 3763. DOI.
- Delgado A, Arco R, Ibarra-Molero B, Sanchez-Ruiz JM. “Using Resurrected Ancestral Proviral Proteins to Engineer Virus Resistance.” Cell Rep, 2017, 19: 1247. DOI.
- Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. “Hyperstability and Substrate Promiscuity in Laboratory Resurrections of Precambrian upbeta-Lactamases.” J Am Chem Soc, 2013, 135: 2899. DOI.
- Zou T, Risso VA, Gavira JA, Sanchez-Ruiz JM, Ozkan SB. “Evolution of Conformational Dynamics Determines the Conversion of a Promiscuous Generalist into a Specialist Enzyme.” Mol Biol Evol, 2014, 32: 132. DOI.
- Risso VA, Martinez-Rodriguez S, Candel AM, Krüger DM, Pantoja-Uceda D, Ortega-Muñoz M, Santoyo-Gonzalez F, Gaucher EA, Kamerlin SCL, Bruix M, Gavira JA, Sanchez-Ruiz JM. “De novo active sites for resurrected Precambrian enzymes.” Nat Commun, 2017, 8: 16113. DOI.
- Gould SJ, Wonderful life: the Burgess shale and the nature of history. W.W. Norton & Company, 1989.
