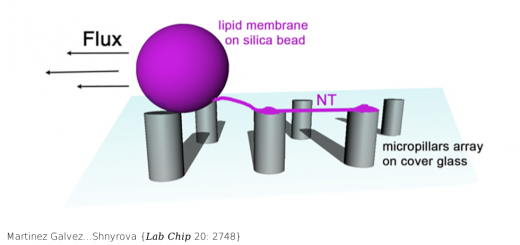
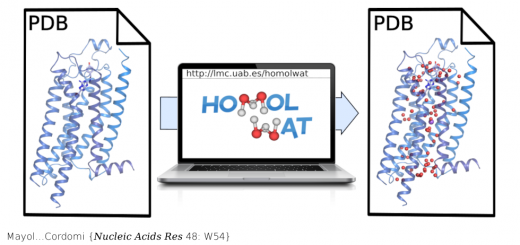
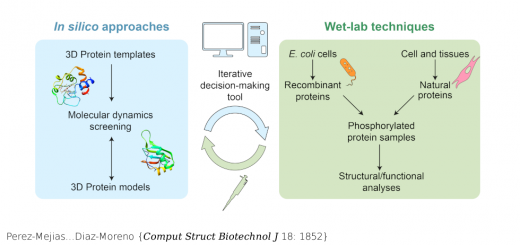
Aggrescan3D (A3D) 2.0: prediction and engineering of protein solubility

A3D 2.0 is an update of the structural aggregation predictor A3D; allowing to model the dynamic aggregation propensity of large multi-chain protein structures like Rubisco -in the figure above- and to automatically improve their solubility.
Kuriata A, Iglesias V, Pujols J, Kurcinski M, Kmiecik S, Ventura S.
Nucleic Acids Res 2019 Jul; 47: W300.
Protein aggregation is a hallmark of a growing number of human disorders and constitutes a major bottleneck in the manufacturing of therapeutic proteins. Therefore, there is a strong need of in-silico methods that can anticipate the aggregative properties of protein variants linked to disease and assist the engineering of soluble protein-based drugs. A few years ago, we developed a method for structure-based prediction of aggregation properties that takes into account the dynamic fluctuations of proteins. The method has been made available as the Aggrescan3D (A3D) web server and applied in numerous studies of protein structure-aggregation relationship. Here, we present a major update of the A3D web server to version 2.0. The new features include: extension of dynamic calculations to significantly larger and multimeric proteins, simultaneous prediction of changes in protein solubility and stability upon mutation, rapid screening for functional protein variants with improved solubility, a REST-ful service to incorporate A3D calculations in automatic pipelines, and a new, enhanced web server interface. A3D 2.0 is freely available at: http://biocomp.chem.uw.edu.pl/A3D2/.
PubMed: 31049593. Doi: 10.1093/nar/gkz321. OPEN Free PMC