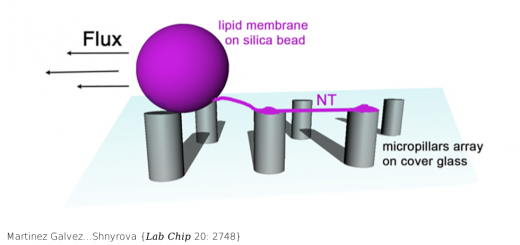
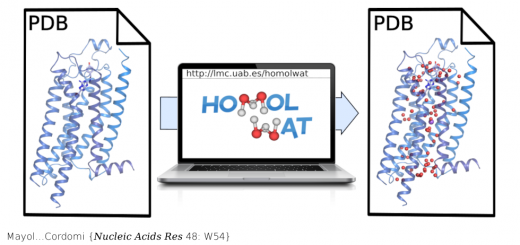
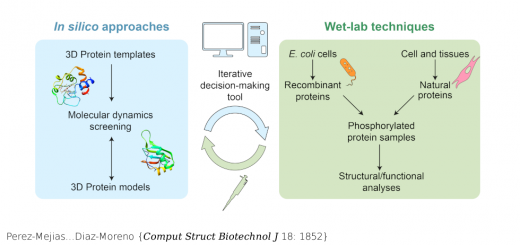
Ortega…Millet {Chem Biol 22: 1597}

Under high salt conditions, live is possible thanks to specific protein adaptations, allowing resistance to extreme osmolarity in the cellular cytosol. In this work we demonstrate that, in presence of ions, the uncommon aminoacid sequence of halophilic proteins favors their native conformation, against the unfolded state, through a synergistic mechanism of salt-induced stability.
Halophilic Protein Adaptation Results from Synergistic Residue-Ion Interactions in the Folded and Unfolded States
Ortega G, Diercks T, Millet O.
Chem Biol 2015 Dec; 22:1597. [Epub 29 Nov. 2015]
Halophilic organisms thrive in environments with extreme salt concentrations and have adapted by allowing molar quantities of cosolutes, mainly KCl, to accumulate in their cytoplasm. To cope with this high intracellular salinity, halophilic organisms modified the chemical composition of their proteins to enrich their surface with acidic and short polar side chains, while lysines and bulky hydrophobic residues got depleted. We have emulated the evolutionary process of haloadaptation with natural and designed halophilic polypeptides and applied novel nuclear magnetic resonance (NMR) methodology to study the different mechanisms contributing to protein haloadaptation at a per residue level. Our analysis of an extensive set of NMR observables, determined over several proteins, allowed us to disentangle the synergistic contributions of protein haloadaptation: cation exclusion and electrostatic repulsion between negatively charged residues destabilize the denatured state ensemble while cumulative weak cation-protein interactions stabilize the folded conformations.
PubMed: 26628359. Doi: 10.1016/j.chembiol.2015.10.010