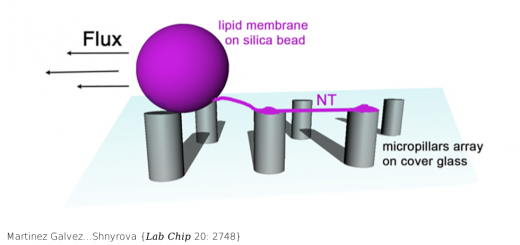
Derivation of nearest-neighbor DNA parameters in magnesium from single molecule experiments
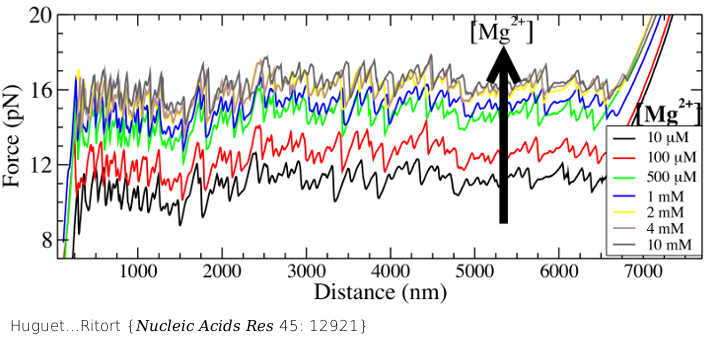
Experimental force-distance curves measured by unzipping DNA by pulling the two strands apart with optical tweezers in aqueous buffers at different divalent salt (magnesium chloride) concentrations. We find that the higher the salt concentration the higher the average unzipping force.
Huguet JM, Ribezzi-Crivellari M, Bizarro CV, Ritort F.
Nucleic Acids Res 2017 Dec; 45: 12921.
DNA hybridization is an essential molecular reaction in biology with many applications. The nearest-neighbor (NN) model for nucleic acids predicts DNA thermodynamics using energy values for the different base pair motifs. These values have been derived from melting experiments in monovalent and divalent salt and applied to predict melting temperatures of oligos within a few degrees. However, an improved determination of the NN energy values and their salt dependencies in magnesium is still needed for current biotechnological applications seeking high selectivity in the hybridization of synthetic DNAs. We developed a methodology based on single molecule unzipping experiments to derive accurate NN energy values and initiation factors for DNA. A new set of values in magnesium is derived, which reproduces unzipping data and improves melting temperature predictions for all available oligo lengths, in a range of temperature and salt conditions where correlation effects between the magnesium bound ions are weak. The NN salt correction parameters are shown to correlate to the GC content of the NN motifs. Our study shows the power of single-molecule force spectroscopy assays to unravel novel features of nucleic acids such as sequence-dependent salt corrections.
PubMed: 29177444. Doi: 10.1093/nar/gkx1161.