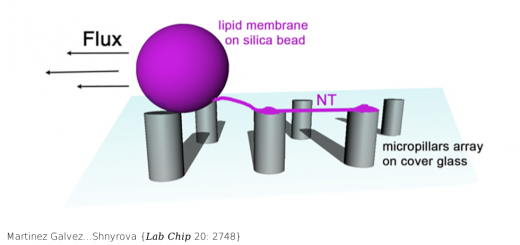
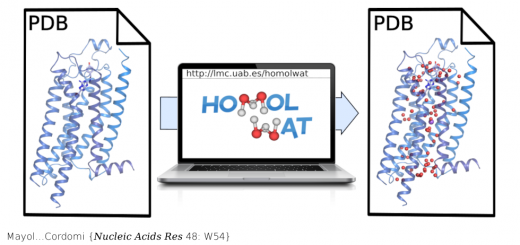
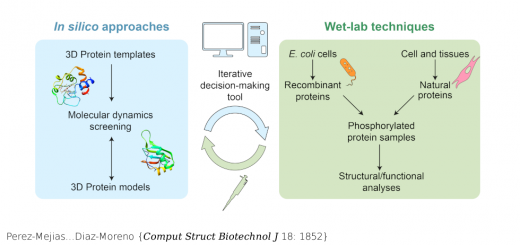
A multi-modal coarse grained model of DNA flexibility mappable to the atomistic level

MC-eNN DNA, a new coarse-grained model for the study of the structure and dynamics of DNA molecules. The model is based on highly accurate atomistic simulations and uses an efficient and very fast sampling method to produce thousands of conformations in a few minutes with low computational requirements.
Walther J, Dans PD, Balaceanu A, Hospital A, Bayarri G, Orozco M.
Nucleic Acids Res 2020 Mar; 48: e29.
We present a new coarse grained method for the simulation of duplex DNA. The algorithm uses a generalized multi-harmonic model that can represent any multi-normal distribution of helical parameters, thus avoiding caveats of current mesoscopic models for DNA simulation and representing a breakthrough in the field. The method has been parameterized from accurate parmbsc1 atomistic molecular dynamics simulations of all unique tetranucleotide sequences of DNA embedded in long duplexes and takes advantage of the correlation between helical states and backbone configurations to derive atomistic representations of DNA. The algorithm, which is implemented in a simple web interface and in a standalone package reproduces with high computational efficiency the structural landscape of long segments of DNA untreatable by atomistic molecular dynamics simulations.
PubMed: 31956910. Doi: 10.1093/nar/gkaa015. OPEN Free PMC