Single molecule research: When biology meets physics
 “Take a single DNA molecule and pull from its extremities, while recording the force-extension curve until it gets fully straightened.”
“Take a single DNA molecule and pull from its extremities, while recording the force-extension curve until it gets fully straightened.”
This thought experiment, which was just a dream a few decades ago, has now become standard in many research institutes worldwide. By labeling the ends of a DNA molecule with specific chemical groups (biotin, avidin, digoxigenin), it is possible to tether a single DNA between two surfaces. By moving one surface relative to the other and using one of them as a force sensor, it is now possible to measure the force-extension curve of single biopolymers, from DNA to RNA and proteins.

Figure 1. (Left) Illustration of the experimental optical tweezers setup used for pulling a single DNA molecule. A bead is captured in an optical trap and used to measure the force exerted on the molecule while the other is immobilized at the tip of a micropipette by air suction (bottom of the illustration). The DNA molecule is labeled at both extremities to tether it between two plastic beads. (Right) Force-extension curve of a half-lambda (24kb) DNA molecule showing different regimes: entropic (enthalpic) response below (above) 10pN and overstretching transition at 65pN, where the DNA overextends by approximately 70% of its natural contour length. Overstretching is known to combine a mixture of distinct DNA structural phases: stretched DNA that forms a ladder, melted DNA that forms bubbles and unpeeled DNA at the extremities of the tether.
A revolutionary tool for biophysics
Human history shows that the invention of novel scientific instruments, leading to new observations and phenomena, irreversibly expands our knowledge toward new unexplored venues. Force measuring techniques, such as optical tweezers, have granted scientists access to new phenomena (e.g. the observation of the overstretching transition in DNA, Figure 1). Of foremost importance among them is the direct measurement of the progression of a molecular reaction along a well-defined coordinate, such as the molecular extension. This gives also the possibility to measure kinetics by monitoring the evolution of a molecular reaction at the single molecule level in real time (e.g. protein folding), overcoming the limitations of ensemble or bulk methods where molecular events (e.g. conformational transitions) are asynchronous in time. By detecting forces in the piconewton (pN) range and nanometer (nm) extensions, SME can measure extremely tiny energies, on the order of pN*nm=10-21 Joule. Together with their high time resolution (sub-millisecond in force measurements), single molecule techniques allow scientists to perform the most accurate determination to date of thermodynamics and kinetics of complex molecular reactions.
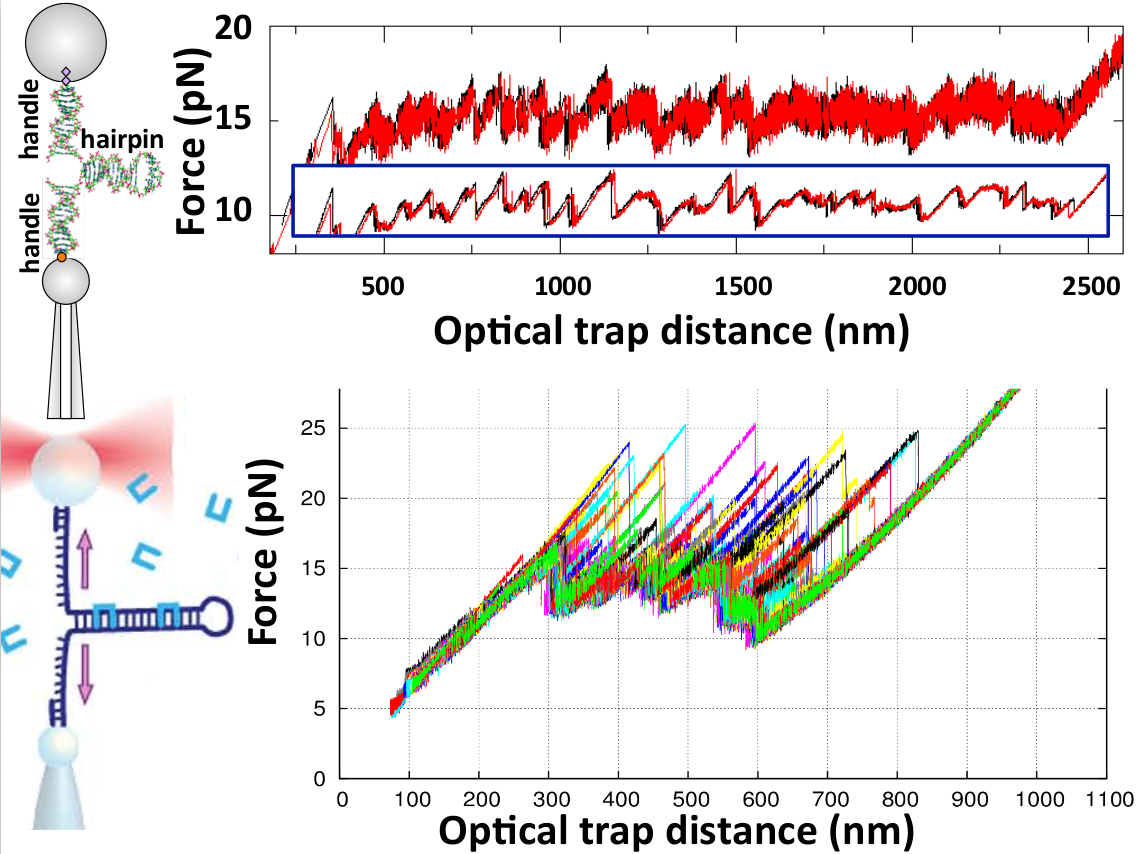
Figure 2. (Upper) Force versus optical-trap position measured in an unzipping (black) and rezipping (red) experiment of a DNA hairpin of 2.2kb at 1kHz acquisition frequency. The force sawtooth pattern at 15pN shows the progressive disruption of the base pairs along the double helix. The last part on the right of the curve corresponds to the elastic response of the single-stranded DNA. Note the strong thermal noise in the curve and the low hysteresis between unzipping and rezipping (black and red curves are practically superimposing). The inset are the same data but filtered to 1Hz bandwith. Data from Ref. [5]. (Lower) Repeated unzipping curves of a 480bp DNA hairpin in the presence of the bis-intercalating peptide Thiocoraline (shown as blue staples in the illustration at the left). Note the large force peaks indicative of DNA binding events. Data from Ref. [6].
The combination of force spectroscopy and fluorescence is quickly expanding the possibilities of SME [7,8]. Simultaneous measurement of forces and efficiency of fluorescence resonance energy transfer (FRET) between donor-acceptor pairs makes it possible to monitor two reaction coordinates at the same time, enhancing the capability of detecting intermolecular binding events or even correlating translocation modes (e.g. elongation, pausing, backtracking) to conformational and allosteric transitions in motor proteins. As a result of these developments, established paradigms in biophysics such as the uniqueness of the native structure in RNAs and proteins are now under dispute, as evidence shows that a multiplicity of native states in enzymes may be involved in a unique biological function.
When signal and noise are equally important
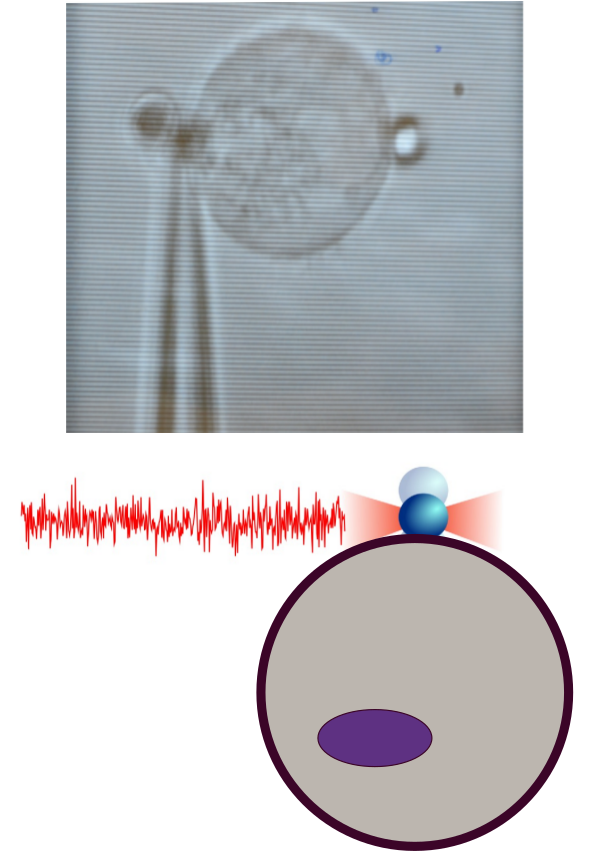
Figure 3. Time-resolved fluctuation spectroscopy. Noise measurements of the force acting on an optically trapped bead bound to the cell membrane (red trace, bottom) it is possible to characterize the spectrum of membrane rigidities of cell populations known as mechanical phenotyping.
Nonequilibrium systems are characterized by the presence of non-zero currents of physically conserved quantities (such as mass, energy, charge, momentum) that, according to the second law of thermodynamics, result into an overall net positive entropy production. However entropy production is positive only when averaged over many experiments or over very long times, whereas thermal fluctuations make the entropy production to be occasionally negative. Fluctuation theorems establish exact symmetry relations between the probability to produce or absorb a given amount of entropy for a given nonequilibrium setting. Such symmetry relations (and the famous Jarzynski equality as a corollary) allow us to recover equilibrium free energy differences from irreversible and noisy work measurements [9]. Ultimately this shows the useful side of the always so annoying noise for biophysical measurements.
When biology meets physics
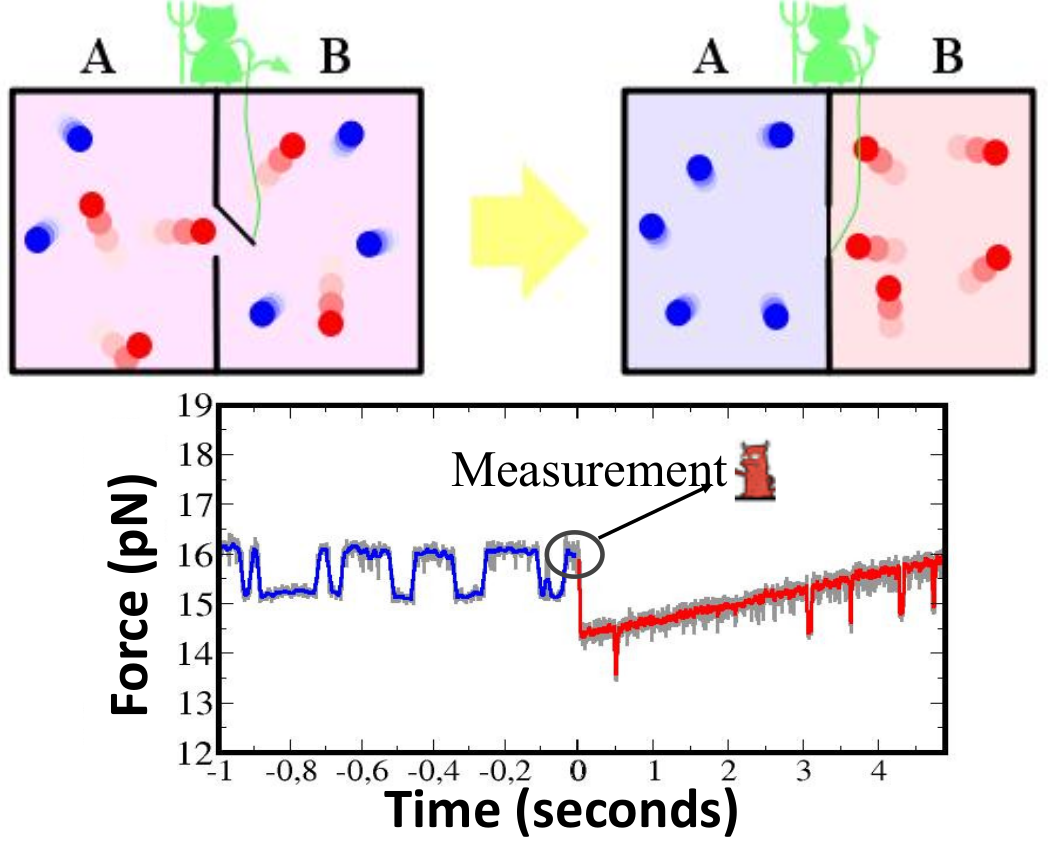
Figure 4. (Upper) The paradox of the Maxwell demon is a thought experiment to violate the second law. A gas vessel contains two compartments separated by a wall with a small gate that can be opened and closed without performing work. A small demon observes the motion of the molecules approaching the gate from each side and closes and opens it to separate fast (red) from slow (blue) molecules. At the end of the process the total entropy has decreased without performing work, against the second law. (Lower) Experimental realization of the Maxwell demon in a single DNA hairpin of 20 bp that hops between the folded and unfolded states. At a given time, a measurement is made and, depending on the molecular state, the force is increased or decreased, according to a predetermined protocol (M. Ribezzi-Crivellari and F. Ritort, unpublished).
Conclusion
SME have emerged as one of the most powerful methodologies to investigate a large variety of biological systems, from single molecules and single cells to the most complex molecular machinery that operates under the concerted action of assembled components. By monitoring the trajectories of individual molecules in space, time and energy, SME gives access to biophysical processes from a new perspective where thermal fluctuations, disorder and information are measurable under generic nonequilibrium conditions. Technological progress going hand by hand with the development of creative biological assays will greatly expand the possibilities of SME in the coming future. Quite probably this will have implications in our understanding of fundamental physical concepts such as energy and information and maybe someday come to understand what is life.
Small Biosystems Lab
Departament de Fisica Fonamental, Facultat de Fisica, Universitat de Barcelona
Diagonal 647, 08028 Barcelona (Spain)
E-mail: ritort@ffn.ub.es
References
- Smith SB, Finzi L, Bustamante C. “Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads”. Science, 1992, 258: 1122. DOI: 10.1126/science.1439819.
- Gonzalez R, Tinoco I. “Biological mechanisms, one molecule at a time”. Genes & Dev, 2011, 25: 1205. DOI: 10.1101/gad.2050011.
- Ritort F. “Single molecule experiments in biological physics: methods and applications”. J Phys Condens Mat, 2006, 18:R531. DOI: 10.1088/0953-8984/18/32/R01.
- Garcia-Parajo MF. “Super-resolution optical nanoscopy gets the 2014 Nobel Prize in Chemistry“. Biofisica, 2015, 1.
- Huguet JM, Bizarro CV, Forns N, Smith SB, Bustamante C, Ritort F. “Single-molecule derivation of salt dependent base-pair free energies in DNA”. Proc Natl Acad Sci, 2010, 107: 15431. DOI: 10.1073/pnas.1001454107.
- Camunas-Soler J, Manosas M, Frutos S, Tulla-Puche J, Albericio F and Ritort F. “Single-molecule kinetics and footprinting of DNA bis-intercalation: the paradigmatic case of Thiocoraline”. Nucleic Acids Res, 2015, 43: 2767. DOI: 10.1093/nar/gkv087.
- Comstock MJ, Ha T, Chemla, YR. “Ultrahigh-resolution optical trap with single-fluorophore sensitivity”. Nat Methods, 2011, 8: 335. DOI: 10.1038/nmeth.1574.
- Heller I, Sitters G, Broekmans OD, Farge G, Menges C, Wende W, Hell SW, Peterman EJG, Wuite, GJL. “STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA”. Nat Methods, 2013, 10: 910. DOI: 10.1038/nmeth.2599.
- Alemany A , Ribezzi-Crivellari M, Ritort F. “From free energy measurements to thermodynamic inference in nonequilibrium small systems”. New J Phy, 2015, 17: 075009. DOI: 10.1088/1367-2630/17/7/075009.
- Koski JV, Maisi VF, Pekola JP, Averin DV. “Experimental realization of a Szilard engine with a single electron”. Proc Natl Acad Sci, 2014, 111: 13786. DOI: 10.1073/pnas.1406966111.
- Dieterich E, Camuñas-Soler J, Ribezzi-Crivellari M, Seifert U, Ritort F. “Single-molecule measurement of the effective temperature in non-equilibrium steady states”. Nat Phys, 2015, 11: 971. DOI: 10.1038/nphys3435.
